Course Description
Theory and Practice of Next Generation Sequencing Analysis
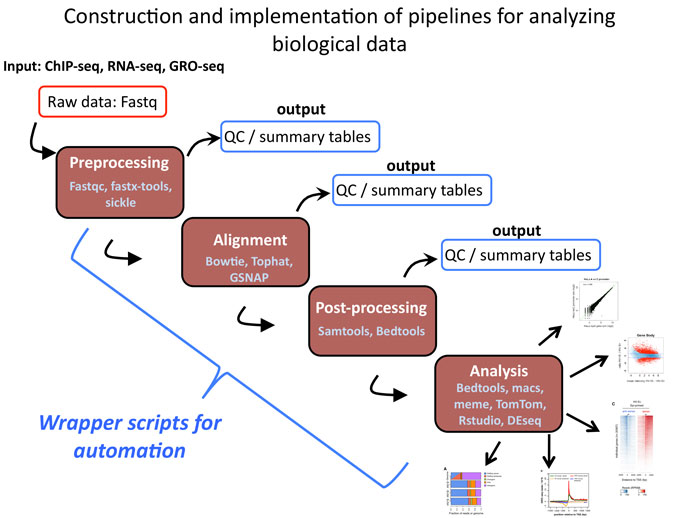
This course will cover the creation of workflows for the processing and analysis of data from next generation sequencing experiments. The focus will be on eukaryotic functional genomics datasets such as ChIP-seq, RNA-seq and possibly GRO-seq. Students will learn basic programming skills necessary to complete these tasks including: commands for navigating and operating in the terminal environment, basic shell scripting for creating pipelines, parsing, and analyzing data. Use of available ‘off the shelf’ analysis tools will also be covered and incorporated into workflows. Students will also be introduced to the R programming language for analysis and display of processed data. After completing this course, student will not only have the skills needed for basic analysis of NGS data, but will have the confidence to begin new or more complex analysis on their own.
Find the complete description here.
Course Workflow
Syllabus